As popular as NoSQL databases currently are, there is still an immense amount
of data in the world for which the relational database is still the most appropriate
way to store, access and manipulate it. For those of us who like to use Python
rather than JDBC, we are fortunate to have bindings for the major databases
which implement the Python Database API, aka PEP249.
This standard is important because it allows you to write your application in a way
that promotes cross-platform (cross-database engine) portability. While you do still
need to be aware of differences between the databases (for example, Oracle's VARCHAR2
vs PostgreSQL's VARCHAR), the essential tasks that you need to accomplish when
making use of a database are abstracted from you.
In this brief introduction I will show you how to install two popular Python database
binding packages, connect to a database and run queries.
Getting a connection to the database
Now that you have the correct packages installed, you can start investigating. The first
thing to do is start an interactive interpreter and import the modules you need:
$ python3.8
>> import cx_Oracle
(Note that unless you're using Solaris' pre-packaged version, you must point LD_LIBRARY_PATH to where
the module can locate the Instant Client libraries, specifically libclntsh.so)
$ python 3.8
>>> import psycopg2
>>> from psycopg2.extras import execute_batch
We need a connection to the database, which in Oracle terms is a DSN, or Data Source Name.
This is made up of the username, password, host (either address or IP), port and database instance name.
Put together, the host, port and database instance are what Oracle calls the "service name".
The common example you will see in Oracle documentation (and on stackoverflow!) is
scott/tiger@orcl:1521/orcl
I'm a bit over seeing that, so I've created a different username and DBname:
DEMOUSER/DemoDbUser1@dbhost:1521/demodb
Since Oracle defaults to using port 1521 on the host, you only need to specify the port number if your
database is listening on a different port.
An Oracle connection is therefore
user = "DEMOUSER"
passwd = "DemoDbUser1"
servicename = "dbhost:1521/demodb"
try:
connection = cx_Oracle.connect(user, passwd, servicename)
except cx_Oracle.DatabaseError as dbe:
print("""Unable to obtain a connection to {servicename}""".format(servicename=servicename))
raise
With PostgreSQL we can also specify whether we want SSL enabled.
try:
connection = psycopg2.connect(dbname=dbname,
user=dbuser,
password=dbpassword,
host=dbhost,
port=dbport,
sslmode=dbsslmode)
except psycopg2.OperationalError as dboe:
print("""Unable to obtain a connection to the database.\n
Please check your database connection details\n
Notify dbadmin@{dbhost}""".format(dbhost=dbhost),
file=sys.stderr, flush=True)
raise
Once we have the connection, we need a cursor so we can execute statements:
cursor = connection.cursor()
Now that we have our connection and a cursor to use, what can we do with it? SQL baby!
We need a handy dataset to muck around with, so I've got just the thing courtesy of
my previous post about determining your electorate. My JSON files have a fairly
basic structure reflecting my needs for that project, but that is still sufficient
our purposes here.
>>> import json
>>> actf = json.load(open("find-my-electorate/json/ACT.json", "r"))
>>> actf.keys()
dict_keys(['Brindabella', 'Ginninderra', 'Kurrajong', 'Murrumbidgee', 'Yerrabi'])
>>> actf["Yerrabi"].keys()
dict_keys(['jurisdiction', 'locality', 'blocks', 'coords'])
The 'jurisdiction' key is the state or territory name (ACT, Australian Capital Territory in this case),
the 'locality' is the electorate name, 'blocks' is a list of the Australian Bureau of Statistics
Mesh Block which the electorate contains, and 'coords' is a list of the latitude, longitude pairs
which are the boundaries of those blocks.
Now we need a schema to operate within. This is reasonably simple - if you've done any work with
SQL databases. To start with, we need two SEQUENCE s, which are database objects from which
any database user may generate a unique integer. This is the easiest and cheapest way that I know
of to generate values for primary key columns. (For a good explanation on them, see the
Oracle 19c CREATE SEQUENCE page). After that, we'll create two tables: electorate and geopoints.
(We're going to ignore the Mesh Block data, it's not useful for this example).
-- This is the Oracle DB form
DROP TABLE GEOPOINTS CASCADE CONSTRAINTS;
DROP TABLE ELECTORATES CASCADE CONSTRAINTS;
DROP SEQUENCE ELECTORATE_SEQ CASCADE;
DROP SEQUENCE LATLONG_SEQ CASCADE;
CREATE SEQUENCE ELECTORATE_SEQ INCREMENT BY 1 MINVALUE 1 NOMAXVALUE;
CREATE SEQUENCE LATLONG_SEQ INCREMENT BY 1 MINVALUE 1 NOMAXVALUE;
CREATE TABLE ELECTORATES (
ELECTORATE_PK NUMBER DEFAULT DEMOUSER.ELECTORATE_SEQ.NEXTVAL NOT NULL PRIMARY KEY,
ELECTORATE VARCHAR2(64) NOT NULL,
STATENAME VARCHAR2(3) NOT NULL -- Using the abbreviated form
);
CREATE TABLE GEOPOINTS (
LATLONG_PK NUMBER DEFAULT DEMOUSER.LATLONG_SEQ.NEXTVAL NOT NULL PRIMARY KEY,
LATITUDE NUMBER NOT NULL,
LONGITUDE NUMBER NOT NULL,
ELECTORATE_FK NUMBER REFERENCES ELECTORATES(ELECTORATE_PK)
);
-- Now for the PostgreSQL form
DROP TABLE IF EXISTS GEOPOINTS CASCADE;
DROP TABLE IF EXISTS ELECTORATES CASCADE;
DROP SEQUENCE IF EXISTS ELECTORATE_SEQ CASCADE;
DROP SEQUENCE IF EXISTS LATLONG_SEQ CASCADE;
CREATE SEQUENCE ELECTORATE_SEQ INCREMENT BY 1 MINVALUE 1 NO MAXVALUE;
CREATE SEQUENCE LATLONG_SEQ INCREMENT BY 1 MINVALUE 1 NO MAXVALUE;
CREATE TABLE ELECTORATES (
ELECTORATE_PK INTEGER DEFAULT NEXTVAL('ELECTORATE_SEQ') NOT NULL PRIMARY KEY,
ELECTORATE VARCHAR(64) NOT NULL,
STATENAME VARCHAR(3) NOT NULL -- Using the abbreviated form
);
CREATE TABLE GEOPOINTS (
LATLONG_PK INTEGER DEFAULT NEXTVAL('LATLONG_SEQ') NOT NULL PRIMARY KEY,
LATITUDE NUMERIC NOT NULL,
LONGITUDE NUMERIC NOT NULL,
ELECTORATE_FK INTEGER REFERENCES ELECTORATES(ELECTORATE_PK) NOT NULL
);
We need to execute these statements one by one, using cursor.execute(). Come back once you've
done that and we've got our schema set up.
[I see that a short time has passed - welcome back]
Now we need to populate our database. For both connection types, we'll make use of prepared
statements, which allow us to insert, update, delete or select many rows at a time. For the
Oracle connection we'll use the executemany() function, but for PostgreSQL's psycopg2
bindings we'll use execute_batch() instead. (See the psycopg2 website note about it).
For the Oracle examples, we'll use a bind variable so that when we INSERT into
the ELECTORATES table we get the ELECTORATE_PK to use in the subsequent INSERT to the
GEOPOINTS table. That saves us a query to get that information. The psycopg2 module
does not have this support, unfortunately. Since we have many records to process, I've created
some small functions to enable DRY
# Oracle version
>>> epk = cursor.var(int)
>>> electStmt = """INSERT INTO ELECTORATES (ELECTORATE, STATENAME)
... VALUES (:electorate, :statename)
... RETURNING ELECTORATE_PK INTO :epk"""
>>> geoStmt = """INSERT INTO GEOPOINTS(LATITUDE, LONGITUDE, ELECTORATE_FK)
... VALUES (:lat, :long, :fk)"""
>>> def addstate(statename):
... for electorate in statename:
... edict = {"electorate": statename[electorate]["locality"],
... "statename": statename[electorate]["jurisdiction"],
... "epk": epk}
... cursor.execute(electStmt, edict)
... points = list()
... for latlong in statename[electorate]["coords"]:
... points.append({"latitude": float(latlong[1]),
... "longitude": float(latlong[0]),
... "fk": int(epk.getvalue(0)[0])})
... cursor.executemany(geoStmt, points)
... connection.commit()
>>> allpoints = dict()
>>> states = ["act", "nsw", "nt", "qld", "sa", "tas", "wa", "vic", "federal"]
>>> for st in states:
... allpoints[st] = json.load(open("json/{stu}.json".format(stu=st.upper()), "r"))
... addstate(allpoints[st])
For the PostgreSQL version, we need to subtly change the INSERT statements:
>>> electStmt = """INSERT INTO ELECTORATES (ELECTORATE, STATENAME)
... VALUES (%(electorate)s, %(statename)s)
... RETURNING ELECTORATE_PK"""
>>> geoStmt = """INSERT INTO GEOPOINTS(LATITUDE, LONGITUDE, ELECTORATE_FK)
... VALUES (%(latitude)s, %(longitude)s, %(fk)s)"""
Another thing we need to change is the first execute, because bind variables are an
Oracle extension, and we're also going to change from executemany() to execute_batch():
>>> def addstate(statename):
... for electorate in statename:
... edict = {"electorate": statename[electorate]["locality"],
... "statename": statename[electorate]["jurisdiction"]}
... cursor.execute(electStmt, edict)
... epk = cursor.fetchone()[0]
... points = list()
... for latlong in statename[electorate]["coords"]:
... points.append({"latitude": float(latlong[1]),
... "longitude": float(latlong[0]), "fk": epk})
... execute_batch(cursor, geoStmt, points)
... connection.commit()
The rest of the data loading is the same as with Oracle. Since I'm curious about
efficiencies, I did a rough benchmark of the data load with executemany() as well,
and it was around half the speed of using execute_batch(). YMMV, of course, so always
test your code with your data and as close to real-world conditions as possible.
Now that we have the data loaded we can do some investigations. I'm going to show the
PostgreSQL output for this and call out differences with Oracle where necessary.
While some parts of our state and territory borders follow rivers and mountain ranges, quite a lot of
them follow specific latitudes and longitudes. The border between Queensland and New South Wales, for
example is mostly along the 29th parallel. Between the Northern Territory and South Australia it's the
26th parallel, and that between South Australia and Western Australia is along meridian 129 East.
If you go to a mapping service and plug in 29S,141E (the line between Queensland and New South Wales):
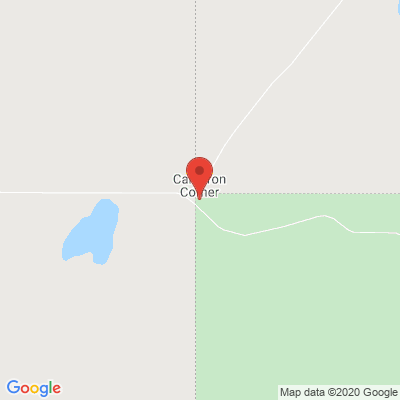
you'll see that the exact point is not where the boundary is actually drawn. That means we need to use
some fuzziness in our matching.
>>> cursor.execute("""SELECT STATENAME, ELECTORATE FROM ELECTORATES E WHERE ELECTORATE_PK IN
... (SELECT DISTINCT ELECTORATE_FK FROM GEOPOINTS WHERE LATITUDE BETWEEN -29.001 AND -28.995)
... ORDER BY STATENAME, ELECTORATE""")
>>> results = cursor.fetchall()
>>> for s, e in results:
... print("{s:6} {e}".format(s=s, e=e))
...
NSW Ballina
NSW Barwon
NSW Clarence
NSW Lismore
NSW New England
NSW Northern Tablelands
NSW Page
NSW Parkes
QLD Maranoa
QLD Southern Downs
QLD Warrego
SA Giles
SA Grey
SA Stuart
SA Stuart
WA Durack
WA Geraldton
WA Kalgoorlie
WA Moore
WA North West Central
Hmm. Not only did I not want SA or WA electorates returned, the database has also given me the federal electorates as well.
Let's update our electorates table to reflect that jurisdictional issue:
>>> cursor.execute("""ALTER TABLE ELECTORATES ADD FEDERAL BOOLEAN""")
>>> connection.commit()
Popping into a psql session for a moment, let's see what we have:
demodb=> \d electorates
Table "public.electorates"
Column | Type | Collation | Nullable | Default
---------------+-----------------------+-----------+----------+-------------------------------------
electorate_pk | integer | | not null | nextval('electorate_seq'::regclass)
electorate | character varying(64) | | not null |
statename | character varying(3) | | not null |
federal | boolean | | |
Indexes:
"electorates_pkey" PRIMARY KEY, btree (electorate_pk)
Referenced by:
TABLE "geopoints" CONSTRAINT "geopoints_electorate_fk_fkey" FOREIGN KEY (electorate_fk) REFERENCES electorates(electorate_pk)
Now let's populate that column. Unfortunately, though, some state electorates have the same name as federal electorates - and
some electorate names exist in more than one state, too! (I'm looking at you, Bass!). Tempting as it is to zorch our db and
start from scratch, I'm going to take advantage of this information:
We added the federal electorate list after all the states,
the federal list was constructed starting with the ACT, and therefore
Federal electorates will have a higher primary key value than all the states and territories.
With that in mind here's the first federal electorate entry:
demodb=> SELECT ELECTORATE_PK, ELECTORATE, STATENAME FROM ELECTORATES WHERE STATENAME = 'ACT' ORDER BY ELECTORATE_PK, STATENAME;
electorate_pk | electorate | statename
---------------+--------------+-----------
1 | Brindabella | ACT
2 | Ginninderra | ACT
3 | Kurrajong | ACT
4 | Murrumbidgee | ACT
5 | Yerrabi | ACT
416 | Bean | ACT
417 | Canberra | ACT
418 | Fenner | ACT
(8 rows)
Let's check how many electorates have a primary key higher than Bean:
>>> cursor.execute("""SELECT COUNT(ELECTORATE_PK), MAX(ELECTORATE_PK) FROM ELECTORATES""")
>>> cursor.fetchall()
[(566, 566)]
And a quick check to see that we do in fact have 151 electorates with that condition:
>>> 566 - 415
151
We do. Onwards.
>>> cursor.execute("""UPDATE ELECTORATES SET FEDERAL = TRUE WHERE ELECTORATE_PK > 415""")
>>> connection.commit()
>>> cursor.execute("""SELECT COUNT(*) FROM ELECTORATES WHERE FEDERAL IS TRUE""")
>>> cursor.fetchall()
[(151,)]
Likewise, we'll set the others to federal=False:
>>> cursor.execute("""UPDATE ELECTORATES SET FEDERAL = FALSE WHERE ELECTORATE_PK < 416""")
>>> connection.commit()
Back to our queries. I want to see both sorts of electorates, but grouped by whether they are federal or
state electorates:
>>> cursor.execute("""SELECT E.STATENAME, E.ELECTORATE, E.FEDERAL FROM ELECTORATES E
... WHERE E.ELECTORATE_PK IN
... (SELECT DISTINCT ELECTORATE_FK FROM GEOPOINTS WHERE LATITUDE BETWEEN -29.001 AND -28.995)
... GROUP BY E.STATENAME, E.ELECTORATE, E.FEDERAL
... ORDER BY STATENAME, ELECTORATE""")
>>> fedstate = cursor.fetchall()
>>> fedstate
[('NSW', 'Ballina', False), ('NSW', 'Barwon', False), ('NSW', 'Clarence', False), ('NSW', 'Lismore', False), ('NSW', 'New England', True), ('NSW', 'Northern Tablelands', False), ('NSW', 'Page', True), ('NSW', 'Parkes', True), ('QLD', 'Maranoa', True), ('QLD', 'Southern Downs', False), ('QLD', 'Warrego', False), ('SA', 'Giles', False), ('SA', 'Grey', True), ('SA', 'Stuart', False), ('WA', 'Durack', True), ('WA', 'Geraldton', False), ('WA', 'Kalgoorlie', False), ('WA', 'Moore', False), ('WA', 'North West Central', False)]
>>> def tfy(input):
... if input:
... return "yes"
... else:
... return ""
>>> for res in fedstate:
... fmtstr = """{statename:6} {electorate:30} {federal}"""
... print(fmtstr.format(statename=res[0], electorate=res[1], federal=tfy(res[2])))
...
NSW Ballina
NSW Barwon
NSW Clarence
NSW Lismore
NSW New England yes
NSW Northern Tablelands
NSW Page yes
NSW Parkes yes
QLD Maranoa yes
QLD Southern Downs
QLD Warrego
SA Giles
SA Grey yes
SA Stuart
WA Durack yes
WA Geraldton
WA Kalgoorlie
WA Moore
WA North West Central
I could have added a WHERE E.FEDERAL = TRUE to the query, too.
Finally, let's see what state electorates in WA and SA are on the border:
>>> cursor.execute("""SELECT STATENAME, ELECTORATE FROM ELECTORATES E
... WHERE ELECTORATE_PK IN
... (SELECT DISTINCT ELECTORATE_FK FROM GEOPOINTS WHERE LATITUDE BETWEEN -60.00 AND -25.995
AND LONGITUDE BETWEEN 128.995 AND 129.1)
... AND FEDERAL = FALSE ORDER BY STATENAME, ELECTORATE""")
>>> results = cursor.fetchall()
>>> for _res in results:
... print("""{statename:6} {electorate}""".format(statename=_res[0], electorate=_res[1]))
...
NT Namatjira
SA Giles
WA Kalgoorlie
WA North West Central
Why do we have that electorate from the Northern Territory? It's because the coordinates are a bit fuzzy and we had to use a range (the BETWEEN) in our query.